Visualization Critique
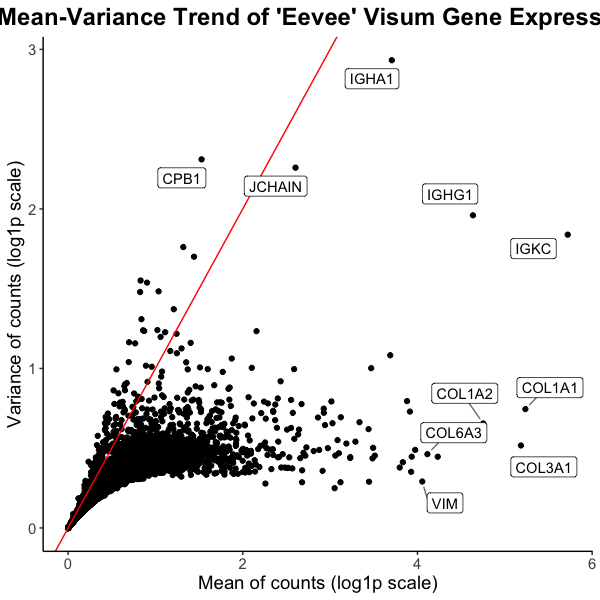
Caleb’s (challin1) visualization seeks to make more salient which genes have high mean or variance in expression. I believe this visualization is very effective because it uses position to encode quantitative data and utilizes the Gestalt principle of continuity to show a trend of points that have similar mean and variance. I believe it could be made more clear which genes have significantly high mean or variance, which could be done using some type of enclosure
# import libraries
library(here)
library(ggplot2)
library(tidyverse)
library(ggrepel)
# read in data
data = read.csv(here("eevee.csv.gz"), row.names = 1)
# QC
dim(data)
head(data)
colnames(data)
rownames(data)
# grab only genes and expression
X = data |>
select(!c(barcode, aligned_x, aligned_y))
# log1p transform data
X = log1p(X)
# get mean and variance for each gene
mean_genes <- apply(X, 2, mean)
var_genes <- apply(X, 2, var)
# combine mean and variance into a data frame
mean_var_df <- data.frame(mean_genes, var_genes)
# add gene names as a column
mean_var_df$genes <- rownames(mean_var_df)
rownames(mean_var_df) <- NULL
# set seed for ggrepel
set.seed(44)
# plot
ggplot(mean_var_df) +
# geom point for mean/variance
geom_point(aes(x = mean_genes, y = var_genes)) +
geom_abline(intercept = 0, slope = 1, color = "red") +
# label high variance genes
geom_label_repel(aes(x = mean_genes, y = var_genes, label=ifelse(var_genes>2, as.character(genes),'')),
box.padding = 0.35,
point.padding = 0.5,
segment.color = 'grey50') +
# label high mean genes
geom_label_repel(aes(x = mean_genes, y = var_genes, label=ifelse(mean_genes>4, as.character(genes),'')),
box.padding = 0.5,
point.padding = 0.5,
segment.color = 'grey50') +
# theme
theme_classic() +
# labels
labs(
x = "Mean of counts (log1p scale)",
y = "Variance of counts (log1p scale)",
title = "Mean-Variance Trend of 'Eevee' Visum Gene Expression"
) +
# text edit
theme(
plot.title = element_text(hjust = 0.5, face="bold", size=18),
text = element_text(size = 14)
)
